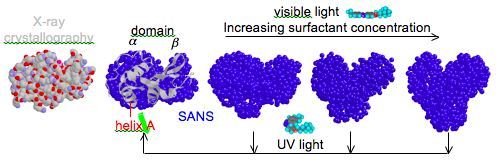
Proteins are the workhorses of biology. Beginning as chains of specific combinations of amino acids, proteins fold due to these amino acids having differing degrees of affinity or aversion for water. It is this seemingly-simple folding event and the resulting shape that proteins adopt in solution, often called the native state, which gives rise to a functional protein, as is the case for most enzymes. However, enzymes are not static entities as suggested by the textbook lock-and-key mechanism. In fact, enzymes regularly undergo conformational changes to intermediately-folded states during the course of activity, particularly upon the interaction with other molecules or as they traverse the activation energy barrier. Thus, information on the native state alone will not provide complete understanding of the function of an enzyme. This instead requires precise knowledge of both the structure of the partially-folded states that form in response to various stimuli, along with the dynamics of the conformational fluctuations of these relatively-flexible partially-folded states. Additionally, selective control of the structure and dynamics of an enzyme may provide similar control over the activity of the enzyme.
We have utilized photoresponsive surfactants as a means to control enzyme activity with simple light illumination. Examples will include (1) photoreversible denaturation and hence deactivation of the enzyme (i.e., a light-triggered on-off switch for the enzymatic reaction), (2) photoreversible partial unfolding to enhance enzyme flexibility and give rise to “superactivity” (i.e., activity greater than the native state), and (3) photoreversible dissociation of enzyme oligomers or enzyme-inhibitor complexes to provide access to the active site (i.e., enzyme reactivation). To thoroughly understand enzyme function during these processes we use small-angle neutron scattering measurements to study the structure of non-native enzyme conformations that form in response to photosurfactant and light. Neutron spin echo measurements further allow examination of the dynamics of conformational flexibility in the partially-folded states relative to the native conformation.
Selected Publications:
1. Wang, S.-C., Mirarefi, P., Faraone, A. & Lee, C.T. “Light-Controlled Protein Dynamics Observed with Neutron Spin Echo Measurements”. Biochemistry 2011, 50, 8150-8162
2. Mirarefi, P.; Lee, C.T. “Photo-induced unfolding and inactivation of bovine carbonic anhydrase in the presence of a photoresponsive surfactant” Biochim. Biophys. Acta 2010, 1804, 106–114
3. Wang, S.-C.; Lee, C.T. “Enhanced Enzymatic Activity through Photoreversible Conformational Changes” Biochemistry 2007, 46, 14557
4. Hamill, A.C; Wang, S.-C.; Lee, C.T. “Probing Lysozyme Conformation with Light Reveals a New Folding Intermediate” Biochemistry 2005, 44, 15139